The same way that bacteria protect their genetic material and cope with stress is now used in a broad range of emerging applications in synthetic biology. By Oscar Blackwell
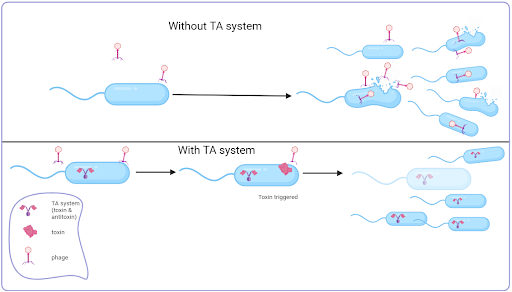
Toxin-antitoxin (TA) systems are prevalent in most prokaryotes. Despite their diversity, most consist of two nearby genetic elements – a gene that codes for a protein toxin and a gene that codes for the counteracting antitoxin, which could also be a protein or an RNA.
One of the first TA systems described was the Hok/Sok system in the 1980s. Its function ensures the maintenance of plasmids in a population. Early experiments demonstrated that insertion of the genes for this TA system into different plasmid types improved levels of plasmid maintenance up to one hundred-fold. The mechanism by which this works is simple and effective. In a cell containing the plasmid, both genes are present and are transcribed; the Sok RNA (the antitoxin) neutralises the Hok RNA (toxin) by binding to it and forming an RNA duplex that can be targeted for enzymatic destruction by the enzyme RNase III. If following cell division, a cell fails to keep the plasmid, the toxin’s RNA persists due to its longer half-life and is now free to be translated to form the toxin, leading to the cell’s death. In this way, the plasmid is retained in the population since any cell without it dies. In this example, the Hok toxin induces cell death by depolarising the membrane. Other toxins work by affecting DNA replication or translation.
Natural plasmids are small bacterial DNA molecules and they can be engineered to carry specific genes or traits. Ensuring plasmid stability is a chief concern when manufacturing proteins in bacterial hosts on a large scale such as in industrial fermentation where bacteria are engineered to carry plasmids that encode genes responsible for the production of desired enzymes. Regulation of TA systems is used to ensure that the cells carry those genes and to strategically improve the process by controlling cell density and resource usage.
TA systems also show promise as bioengineering tools in DNA cloning. One protocol for identifying novel microbial strains is called the StabyCloning™ system developed by Delphi Genetics, which is based on the CcdA/CcdB TA module. The host cell's CcdA antitoxin is truncated (shortened) and inactivated before the addition of a 14 base pair sequence that fuses to the antitoxin essentially activating the antitoxin which can now neutralise the CcdB toxin.
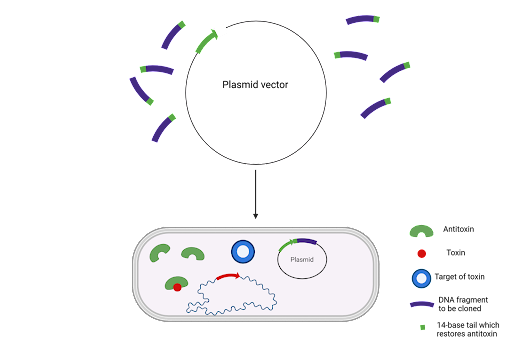
This allows for the positive selection of recombinant plasmids as only cells with the plasmids that incorporate the sequence can survive without antibiotics. The use of antibiotics as an alternative selection marker was, and is, used extensively, but technology is moving away from this method because it increases the risk of allergic reactions and antibiotic resistant pathogenic bacteria. Recently, the Staby method has been used to develop DNA vaccines without antibiotic resistance genes. In a study by Anca Reschner and her team, they created a new plasmid vector which contained the CcdA gene, a promoter for high expression and the specific viral antigen for Aujeszky’s disease. To evaluate the vaccine’s efficacy, mice were challenged with a lethal dose of the virus and those vaccinated with the DNA vaccine showed no signs of the disease. The role of the CcdA gene is crucial to the vaccine’s production process by contributing to the purity (by eliminating plasmid-free cells) and safety (by minimising the risk of contamination with antibiotic resistance genes).
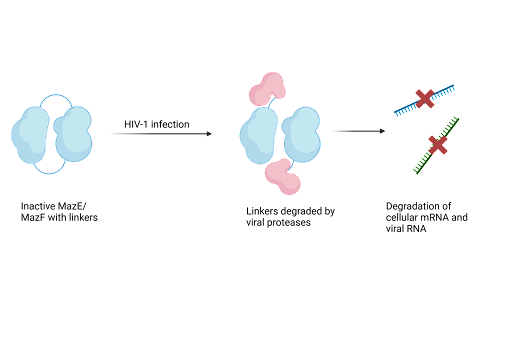
Due to their ability to cause cell death, TA systems have a purpose as antiviral drugs. Recent research has focused on the MazF-MazE TA which can be designed to destroy HIV-infected cells. The system uses the endoribonuclease activity of the MazF toxin to prevent viral infection. Endoribonucleases are a type of enzyme that cut nucleic acids internally. Researchers created a construct consisting of the toxin fused to unstructured MazE, which has been modified so that it serves as a structural component rather than an active antitoxin. The engineered plasmid carrying this MazE-MazF construct is transformed into host cells, providing the system's machinery. Upon HIV-1 infection, viral proteases cleave the linker holding the construct together, thereby releasing the MazF toxin that targets, binds and cleaves cellular mRNA and viral RNA. In this way, viral proliferation is suppressed. As HIV increases, cleavage of the construct also increases to release the active MazF toxin that suppresses HIV.
TA systems’ versatility have shown the world that they provide solutions to global issues across diverse scientific landscapes. Further studies will hopefully unlock their potential to become a prominent tool in the arsenal of combatting antiviral drugs without relying on antibiotic resistance. There are still several potential future realms where they may become useful. For instance, speculation exists about how TA systems could play a role in bioremediation where specific pollutants and contaminants are targeted.
Article written by Oscar Blackwell
References:
Cover photo by CDC on Unsplash
- Jianzhong Lin, Yunxue Guo, Jianyun Yao, Kaihao Tang, Xiaoxue Wang, Applications of toxin-antitoxin systems in synthetic biology, Engineering Microbiology, Volume 3, Issue 2, (2023), 100069, ISSN 2667-3703, https://doi.org/10.1016/j.engmic.2023.100069
- Gerdes, K. The parB (hok/sok) Locus of Plasmid R1: A General Purpose Plasmid Stabilization System. Nat Biotechnol 6, 1402–1405 (1988). https://doi.org/10.1038/nbt1288-1402
- Reschner, A., Scohy, S., Vandermeulen, G., Daukandt, M., Jacques, C., Michel, B., Nauwynck, H., Xhonneux, F., Préat, V., Vanderplasschen, A., & Szpirer, C. (2013). Use of Staby(®) technology for development and production of DNA vaccines free of antibiotic resistance gene. Human vaccines & immunotherapeutics, 9(10), 2203–2210. https://doi.org/10.4161/hv.25086
- Microsoft Word - StabyCloning Tech Note MicrobialDiv (serva.de) , accessed Dec 2023, https://www.serva.de/www_root/documents/StabyCloning%20Tech%20Note%20MicrobialDiv.pdf
- Park, J. H., Yamaguchi, Y., & Inouye, M. (2012). Intramolecular regulation of the sequence-specific mRNA interferase activity of MazF fused to a MazE fragment with a linker cleavable by specific proteases. Applied and environmental microbiology, 78(11), 3794–3799. https://doi.org/10.1128/AEM.00364-12